Students Earn Coauthorship on Published Genomics Research
Work propels students toward promising careers in bioinformatics and genetics.
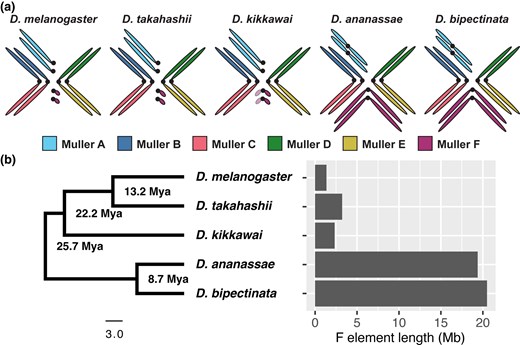
Bennington College undergraduates are now published contributors to a peer-reviewed genomics paper. The paper, which appeared in the journal Molecular Biology and Evolution in December, is titled “Recombination Suppression Drives Expansion of the Drosophila Dot Chromosome.” Through manual gene curation and comparative genomic analysis, students helped identify chromosome-specific size expansion across species of fruit fly.
The research was coordinated through the Genomics Education Partnership (GEP), which includes students and researchers from numerous colleges and universities across the country, including Bennington College. While the partnership itself has produced multiple publications over the years, this is the first time Bennington undergraduates have been included as coauthors.
“The ability to analyze these increasingly available large data sets using bioinformatic tools is an important part of modern biology,” said Bennington College faculty member in Biology Amie McClellan, who joined GEP in 2015.
Genome Jumpstart, designed and taught by McClellan, is a 2000-level course open to students with a wide range of backgrounds, from those new to biology to those planning advanced study in the sciences. The class begins with foundational concepts—how genetic information flows from DNA to RNA to protein—before moving into applied genomic analysis.
“This class was my introduction to bioinformatics,” said Shashvat Shah ’26, one of the students who coauthored the research. “Before the class, I had no clue that biology could be looked at in that manner. It blew my mind that you could look at each gene, divide it into all of its parts, and even look at the exact coordinates where it is located on the chromosome.”
He has since had several internships in the field and is planning to pursue it as a graduate student.
Similarly, Genome Jumpstart was Atlas Seres ’26’s introduction to both bioinformatics and collegiate-level STEM classes.
“Coming to Bennington I wasn't sure what it was exactly that I was interested in, just that I wanted to do something with genetics. I instantly took to gene annotation and grew to love the admittedly tedious work,” Seres said. “Now, almost seven terms later, Genome Jumpstart has heavily influenced the trajectory of my academic plan and post-grad aspirations.”
Genome Jumpstart provides a course-embedded undergraduate research experience, known as a CURE. CUREs are designed to give large numbers of students hands-on experience with authentic scientific research, even if they are not working in a traditional lab setting.
“It’s seen as a more inclusive and equitable way to expose students to the process of scientific research, outside of the resource-intensive environment of a ‘wet lab.’ All it takes to participate is a computer and access to the internet,” McClellan said.
Using an online genome browser developed for the GEP, students analyze segments of fruit fly DNA. While the common fruit fly Drosophila melanogaster has been extensively studied for nearly a century, many related species are far less understood. Students compare predicted gene structures in these lesser-known species to what is known from melanogaster and look for missing genes, rearrangements, or changes in gene orientation.
“These are the types of rearrangements that can happen throughout evolutionary time,” McClellan explained. “By looking at those changes, we can learn how genomes evolve and apply that knowledge to other organisms.”
The work is challenging. Students must master new tools quickly and complete detailed reports explaining and defending their interpretations of the data.
“It was certainly hard at first, but the more time you spend with it, the more intuitive it becomes,” said Shah.
Conflicting evidence is common, and part of the learning process involves revising hypotheses when the data does not fit initial expectations.
“Science is never yes or no,” McClellan said. “It’s ‘maybe this, maybe that,’ and learning how to reconcile those possibilities is part of becoming a scientist. By working on their team projects independently, using their own observations and knowledge to support gene model construction, students realize they are stakeholders in the research.”
“As my first publication, it's certainly an honor,” said Shah. “Knowing that something I worked on is out there for people to look at brings me a lot of joy. It makes me want to keep working for more publications. Through my current internship, I have the opportunity to be involved in a couple more papers and I am looking forward to sharing my work with the research community.”
Seres agreed, “This being my first ever publication, even as a contributor, this is a huge deal. I feel like there is now a certain validity to my claim that I am a scientist. There's tangible evidence, both in the supplementary materials and on the cover of the journal, that I was there and made a contribution."